Sub-groups of space groups
July 3, 2008 Posted by Emre S. Tasci
ISOTROPY is a highly effective program written by the guru of applications of Group Theory on Phase Transitions, Harold T. Stokes (For those who are interested in the topic, I recommend his Introduction to Isotropy Subgroups and Displacive Phase Transitions titled paper(co-authored by Dorian M. Hatch) and Structures and phase transitions in perovskites – a group-theoretical approach paper (co-authored by Christopher J. Howard).
I’m pretty new and noobie in Group Theory – it was one of the subjects I found myself always running away. Anyway, I wanted to limit my search for some specific phase transition search in the database. Say, if an A-B binary has been reported in S1 and S2 structures, I didn’t want to go all the way looking for possible transition mechanisms if S1->S2 isn’t allowed by the group theory at all!
In the meantime, I’ve begun experimenting with the ISOTROPY program, yet I’m still at the very beginning. You can find detailed information and an online version in the related website, but to give an example, suppose we’d like to know about the subgroups of spacegroup 123 (P4/mmm). We enter the following commands:
VALUE PARENT 123
SHOW PARENT
SHOW SUBGROUP
SHOW LATTICE
DISPLAY ISOTROPY
and it displays a list of which a portion is presented below:
Parent Lattice Subgroup Lattice
123 P4/mmm q 123 P4/mmm q
123 P4/mmm q 47 Pmmm o
123 P4/mmm q 83 P4/m q
123 P4/mmm q 65 Cmmm o-b
123 P4/mmm q 10 P2/m m
123 P4/mmm q 12 C2/m m-b
123 P4/mmm q 2 P-1 t
123 P4/mmm q 89 P422 q
123 P4/mmm q 111 P-42m q
123 P4/mmm q 99 P4mm q
123 P4/mmm q 115 P-4m2 q
123 P4/mmm q 25 Pmm2 o
123 P4/mmm q 38 Amm2 o-b
123 P4/mmm q 6 Pm m
123 P4/mmm q 123 P4/mmm q
123 P4/mmm q 127 P4/mbm q
123 P4/mmm q 127 P4/mbm q
You may have noticed that, there are some recurring lines – this is due to some other properties that we have not specifically selected for display, i.e., it would display lines only containing "123 P4/mmm" if we hadn’t specifically asked for subgroup and lattice information (in analogy with the degeneracies in QM).
I needed a table containing all the subgroups of each space-group but there were two problems:
1. As far as I know, ISOTROPY does not accept an input file where you can pass the commands you’ve prepared via a script/macro. So, I was afraid that I would type "VALUE PARENT 1 – DISPLAY ISOTROPY – VALUE PARENT 2 – DISPLAY ISOTROPY – …" (by the way, ISOTROPY has a clever interpreter so that you can just type the first letters of commands and parameters as long as they are not dubious).
2. Again, as far as I know, you can’t direct the output to a file, meaning you have to copy the output manually and paste into a file (x230 in my case).
Luckily, it turned out that:
1. You could not supply an input file, but you could instead, paste the contents of such an input file and ISOTROPY would process it line by line.
2. Stokes had included a very useful and helpful property that all the sessions was recorded automatically in the "iso.log" file.
So, all I had to do was
1. Write a script that would produce the query to retrieve the subgroups of all 230 space-groups.
2. Paste the outcome of this script to ISOTROPY.
3. Write another script that would parse the "iso.log" file and split these reported subgroups per spacegroup (and while I’m at it, remove the duplicate lines and order the subgroups).
Maybe you won’t believe me but I am also not happy to paste lines and lines of code to this blog’s entries. I will deal it soon but in the mean time, please bear with them a little longer.. Ath the moment, I will describe the process and will include them at the end.
* build the query via query_builder.php
* feed this query to ISOTROPY
* split the space groups from the "iso.log" via split_spacegroups.php
* run sortall.php which :
* runs the sortkillduplines.php on each of these files
* sorts again via the sort.pl wrt to the 4th col (space number of the subgroup)
* removes the blank lines
* And that’s it..
Here comes the codes (php+perl)
query_builder.php:
<?
// Generates the query that will be "pasted" into the ISOTROPY prompt
// The query will be stored in isotropy_query.txt
$fp = fopen("isotropy_query.txt","w");
fwrite($fp,"PAGE 1000\nSHOW SUBGROUP\nSHOW LATTICE\nSHOW PARENT\n");
for($i=1;$i<231;$i++)
fwrite($fp,"VALUE PARENT $i\nDISPLAY ISOTROPY\n");
fwrite($fp,"\n\n");
fclose($fp);
?>
sortkillduplines:
<?
// PERL sorts the mentioned file (via $_GET["file"]) and deletes duplicate lines
require("commandline.inc.php");
if($_GET["file"]) $file = $_GET["file"];
else
{
$in = fopen(âphp://stdinâ, ârâ);
$fw = fopen("sortkilldup_tmp.tmp","w");
while(!feof($in))
{
fwrite($fw, rtrim(fgets($in, 4096))."\n");
}
fclose($fw);
$file = "sortkilldup_tmp.tmp";
}
if($_GET["seperator"])$f_seperator = TRUE;//Means that an extra seperator has been placed for this order purpose and is seperated from the text by the parameter defined with seperator=xxx.
if($_GET["n"]||$_GET["numeric"]||$_GET["num"])$f_numeric = TRUE; // If it is TRUE than it is assumed that the first word of each line is a number and will be sort accordingly.
// Reference : http://www.stonehenge.com/merlyn/UnixReview/col06.html
if($f_numeric) $exec = exec("perl -e âprint sort numerically <>;\nsub numerically { \$a <=> \$b; }â ".$file." > sorttemp.txt");
else $exec = exec("perl -e âprint sort <>â ".$file." > sorttemp.txt");
$fi = file("sorttemp.txt");
unlink("sorttemp.txt");
$curlinecont = "wrwerwer";
if(!$f_seperator)
{
for($i=0; $i<sizeof($fi); $i++)
{
$line = rtrim($fi[$i]);
if($curlinecont!=$line)
{
echo $line."\n";
$curlinecont = $line;
}
}
}
else
{
$frs = fopen("sorttemp2.txt","w"); // will capture the dup killed output in this file and resort it later.
for($i=0; $i<sizeof($fi); $i++)
{
$lineorg = rtrim($fi[$i]);
$linearr = explode($_GET["seperator"],$lineorg);
if($curlinecont!=$linearr[0])
{
fwrite($frs, $linearr[1]."\n");
$curlinecont = $linearr[0];
}
}
fclose($frs);
// resort the file
passthru("perl -e âprint sort <>â sorttemp2.txt");
unlink("sorttemp2.txt");
}
// unlink("sortkilldup_tmp.tmp");
?>
split_spacegroups.php:
<?
// splits the isotropy.log file generated by the query :
// PAGE 1000
// SHOW SUBGROUP
// SHOW LATTICE
// SHOW PARENT
// VALUE PARENT 1
// DISPLAY ISOTROPY
// â¦
// VALUE PARENT 230
// DISPLAY ISOTROPY
// with respect to the parent groups
$file = fopen("iso.log", "r") or exit("Unable to open file!");
//Output a line of the file until the end is reached
$cur = 0;
$fp = fopen("dummy.txt","w"); // we need to open a dummy file in order to include the fclose() in the iteration
while(!feof($file))
{
// Read the line
$line = fgets($file);
// Check if it is a spacegroup information entry by looking at the first word
// - if this word is a number, than process that line
if(ereg("^([0-9]+)",$line,$num))
{
$num = $num[0];
if($cur!=$num)
{
// We have a new space group, so create its file:
fclose($fp); // First close the previous space groupâs file
$filename = sprintf("%03d",$num)."_sg.txt";
$fp = fopen($filename,"w");
$cur = $num; // Set the current check data to the sg number
}
fwrite($fp,$line);
}
}
fclose($file);
unlink("dummy.txt");
?>
sortall.php:
<?
// sorts & kills dupes in all the spacegroup files
for($num=1;$num<231;$num++)
{
$filename = sprintf("%03d",$num)."_sg.txt";
$exec = exec("php ../../toolbox/sortkillduplines.php file=".$filename." > temp");
$exec = exec("./sort.pl temp > temp2");
$exec = exec("perl -pi -e \"s/^\\n//\" < temp2 > ".$filename);
}
unlink("temp");
unlink("temp2");
?>
or you can directly skip all these steps and download the generated spacegroup files from here. 8)
Lattice values are given in Schoenflies Notation where it corresponds to (with Pearson notation):
T : triclinic (AP)
M : prmitive monoclinic (MP)
M-B : base-centered monoclinic (MC)
O : primitive orthorhombic (OP)
O-B : base-centered orthorhombic (OC)
O-V : body-centered orthorhombic (OI)
O-F : face-centered orthorhombic (OF)
Q : primitive tetragonal (TP)
Q-V : body-centered tetragonal (TI)
RH : trigonal (HR)
H : hexagonal (HP)
C : primitive cubic (CP)
C-F : face-centered cubic (CF)
C-V : body-centered cubic (CI)
Quick reference on Bravais Lattices and info on how/why they are called after Bravais but not Frankenheim..
The marriage of heaven (common elements) and hell (code writing code).
June 26, 2008 Posted by Emre S. Tasci
So, as Blake had said, "Energy is Eternal Delight." or in other words, let’s merge the preceding two entries to write a code for a function that produces MySQL queries that will fetch the common elements involved with the given elements:
function fetch_elements($el,$order,$total)
{
// produces the query to select the elements that have binaries with the $el
// $order and $total are used to adjust the tab positions and designating the sets
$abc = "ABCDEFGHIJKLMNOPQRSTUVWXYZ";
$tab = str_repeat("\t",($total-$order));
$lom = <<<LOM
$tab SELECT symbol_A AS symb, val1 AS val$abc[$order]
$tab FROM `dbl014`
$tab WHERE symbol_B = "$el"
$tab GROUP BY Symbol_A
$tab UNION
$tab SELECT symbol_B AS symb, val1 AS val$abc[$order]
$tab FROM `dbl014`
$tab WHERE (
$tab symbol_A = "$el"
$tab AND symbol_B != ""
$tab )
$tab GROUP BY Symbol_B
LOM;
return $lom;
}
function merge_queries($str1,$str2,$or1,$or2,$tab)
{
// merges two node (or node group) queries (formed via fetch_elements() function)
// using the INNER JOIN function
return "$tab SELECT *\n$tab FROM (\n".$str1."\n$tab ) AS $or1\n$tab INNER JOIN (\n".$str2."\n$tab ) AS $or2\n$tab USING (symb)\n$tab ";
}
function fetch_common_elements($ar_el)
{
// produces the query that selects the common elements that have binaries with all of the elements given in the $ar_el array (using their symbols)
$total = sizeof($ar_el);
$abc = "ABCDEFGHIJKLMNOPQRSTUVWXYZ";
//$tab = str_repeat("\t",($total-$order-1));
$str[0] = fetch_elements($ar_el[0],0,$total-1);
$str[1] = fetch_elements($ar_el[1],1,$total);
$stri = merge_queries($str[0],$str[1],"A","B",str_repeat("\t",($total-2)));
for($i=2;$i<$total;$i++)
{
$str[$i] = fetch_elements($ar_el[$i],$i,$total);
$stri = merge_queries($str[$i],$stri,$abc[$i],$abc[($i+1)],str_repeat("\t",($total-$i-1)));
}
return $stri;
}
Now, we can call it with, for example:
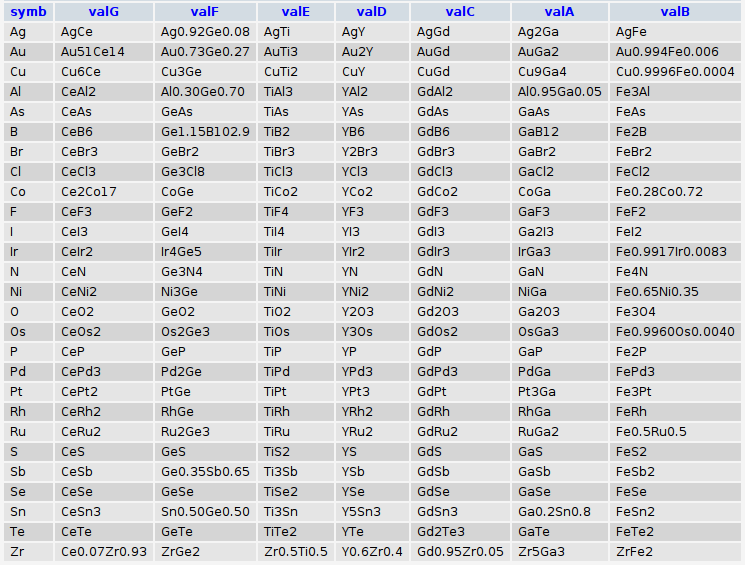
echo fetch_common_elements(Array("Ga","Fe","Gd","Y","Ti","Ge","Ce"));
Which will output:
SELECT *
FROM (
SELECT symbol_A AS symb, val1 AS valG
FROM `dbl014`
WHERE symbol_B = "Ce"
GROUP BY Symbol_A
UNION
SELECT symbol_B AS symb, val1 AS valG
FROM `dbl014`
WHERE (
symbol_A = "Ce"
AND symbol_B != ""
)
GROUP BY Symbol_B
) AS G
INNER JOIN (
SELECT *
FROM (
SELECT symbol_A AS symb, val1 AS valF
FROM `dbl014`
WHERE symbol_B = "Ge"
GROUP BY Symbol_A
UNION
SELECT symbol_B AS symb, val1 AS valF
FROM `dbl014`
WHERE (
symbol_A = "Ge"
AND symbol_B != ""
)
GROUP BY Symbol_B
) AS F
INNER JOIN (
SELECT *
FROM (
SELECT symbol_A AS symb, val1 AS valE
FROM `dbl014`
WHERE symbol_B = "Ti"
GROUP BY Symbol_A
UNION
SELECT symbol_B AS symb, val1 AS valE
FROM `dbl014`
WHERE (
symbol_A = "Ti"
AND symbol_B != ""
)
GROUP BY Symbol_B
) AS E
INNER JOIN (
SELECT *
FROM (
SELECT symbol_A AS symb, val1 AS valD
FROM `dbl014`
WHERE symbol_B = "Y"
GROUP BY Symbol_A
UNION
SELECT symbol_B AS symb, val1 AS valD
FROM `dbl014`
WHERE (
symbol_A = "Y"
AND symbol_B != ""
)
GROUP BY Symbol_B
) AS D
INNER JOIN (
SELECT *
FROM (
SELECT symbol_A AS symb, val1 AS valC
FROM `dbl014`
WHERE symbol_B = "Gd"
GROUP BY Symbol_A
UNION
SELECT symbol_B AS symb, val1 AS valC
FROM `dbl014`
WHERE (
symbol_A = "Gd"
AND symbol_B != ""
)
GROUP BY Symbol_B
) AS C
INNER JOIN (
SELECT *
FROM (
SELECT symbol_A AS symb, val1 AS valA
FROM `dbl014`
WHERE symbol_B = "Ga"
GROUP BY Symbol_A
UNION
SELECT symbol_B AS symb, val1 AS valA
FROM `dbl014`
WHERE (
symbol_A = "Ga"
AND symbol_B != ""
)
GROUP BY Symbol_B
) AS A
INNER JOIN (
SELECT symbol_A AS symb, val1 AS valB
FROM `dbl014`
WHERE symbol_B = "Fe"
GROUP BY Symbol_A
UNION
SELECT symbol_B AS symb, val1 AS valB
FROM `dbl014`
WHERE (
symbol_A = "Fe"
AND symbol_B != ""
)
GROUP BY Symbol_B
) AS B
USING (symb)
) AS D
USING (symb)
) AS E
USING (symb)
) AS F
USING (symb)
) AS G
USING (symb)
) AS H
USING (symb)
and when fed to the database will yield:
thank y’all.. 8)
A code to write code (as in a war to end -all- wars – 8P)
June 19, 2008 Posted by Emre S. Tasci
Lately, I’ve been studying on the nice and efficient algorithm of Dominik Endres and he’s been very helpful in replying to my relentless mails about the algorithm and the software. Now, if you have checked the algorithm, there is a part where iteration is needed to be done over the possible bin boundary distributions. Via a smart workaround he avoids this and uses a recursive equation.
For my needs, I want to follow the brute-force approach. To requote the situation, suppose we have K=4 number of classes, let’s label these as 0,1,2,3. There are 3 different ways to group these classes into 2 (M=2-1=1) bins obeying the following rules:
* There is always at least one classification per bin.
* Bins are adjascent to each other.
* Bins can not overlap.
The number of configurations are given by the formula:
N=(K-1)! / ( (K-M-1)! M!) (still haven’t brought back my latex renderer, sorry for that!)
and these 3 possible configurations for K=4, M=1 are:
|—-|—-|—-|
0 3 : Meaning, the first bin only covers the 0 class while the second bin covers 1, 2, 3.
1 3 : Meaning, the first bin covers 0, 1 while the second bin covers 2, 3.
2 3 : Meaning, the first bin covers 0, 1, 2 while the second bin covers 3.
Now, think that you are writing a code to get this values, it is easy when you pseudo-code it:
for k_0 = 0 : K-1-M
for k_1 = k_0+1 : K-M
…
for k_(K-3) = k_(K-4)+1 : K-4
for k_(K-2) = k_(K-3)+1 : K-3
print("k_0 – k_1 – … – k_(K-2) – K-1")
endfor
endfor
…
endfor
endfor
easier said than done because the number of the k_ variables and the for loops are changing with M.
So after some trying in Octave for recursive loops with the help of the feval() function, I gave up and wrote a PHP code that writes an Octave code and it was really fun! 8)
<?
//Support for command line variable passing:
for($i=1;$i<sizeof($_SERVER["argv"]);$i++)
{
list($var0,$val0) = explode("=",$_SERVER["argv"][$i]);
$_GET[$var0] = $val0;
}
$K = $_GET["K"];
$M = $_GET["M"];
if(!$_GET["K"])$_GET["K"] = 10;
if(!$_GET["M"])$_GET["M"] = 2;
if(!$_GET["f"])$_GET["f"] = "lol.m";
$fp = fopen($_GET["f"],"w");
fwrite($fp, "count=0;\n");
fwrite($fp, str_repeat("\t",$m)."for k0 = 0:".($K-$M-1)."\n");
for($m=1;$m<$M;$m++)
{
fwrite($fp, str_repeat("\t",$m)."for k".$m." = k".($m-1)."+1:".($K-$M-1+$m)."\n");
}
fwrite($fp, str_repeat("\t",$m)."printf(\"".str_repeat("%d – ",$M)."%d\\n\",");
for($i=0;$i<$m;$i++)
fwrite($fp, "k".$i.",");
fwrite($fp,($K-1).")\n");
fwrite($fp, str_repeat("\t",$m)."count++;\n");
for($m=$M-1;$m>=0;$m–)
{
fwrite($fp, str_repeat("\t",$m)."endfor\n");
}
fwrite($fp,"if(count!=factorial(".$K."-1)/(factorial(".$K."-".$M."-1)*factorial(".$M.")))\n\tprintf(\"Houston, we have a problem..\");\nendif\n");
//fwrite($fp,"printf(\"count : %d\\nfactorial : %d\\n\",count,factorial(".$K."-1)/(factorial(".$K."-".$M."-1)*factorial(".$M.")));\n");
fclose($fp);
system("octave ".$_GET["f"]);
?>
Now, if you call this code with for example:
> php genfor.php K=4 M=1
it produces and runs this Octave file lol.m:
count=0;
for k0 = 0:2
printf("%d – %d\n",k0,3)
count++;
endfor
if(count!=factorial(4-1)/(factorial(4-1-1)*factorial(1)))
printf("Houston, we have a problem..");
endif
which outputs:
0 – 3
1 – 3
2 – 3
And you get to keep your hands clean 8)
Later addition (27/6/2008) You can easily modify the code to also give you possible permutations for a set with K items and groups of M items (where ordering is not important). For example, for K=5, say {0,1,2,3,4} and M=4:
0-1-2-3
0-1-2-4
0-1-3-4
0-2-3-4
1-2-3-4
5 (=K!/(M!(K-M)!)) different sets.
The modifications we make to the code are:
* Add an optional parameter that will tell the code that, we want permutation instead of binning:
if($_GET["perm"]||$_GET["p"]||$_GET["P"])
{
$f_perm = TRUE;
$K+=1;
}
via the f_perm flag, we can check the type of the operation anywhere within the code. K is incremented because, previously, we were setting the end point fixed so that the last bin would always include the last point for the bins to cover all the space. But for permutationing we are not bounded by this fixication so, we have an additional freedom.
* Previously, we were just including the last point in the print() statement, we should fix this if we are permutating:
$str = "";
for($i=0;$i<$m;$i++)
$str .= "k".$i.",";
if(!$f_perm) fwrite($fp,$str.($K-1).")\n");
else fwrite($fp, substr($str,0,-1).")\n");
The reason for instead of now writing directly to the file in the iteration we just store it in a variable (str) is to deal with the trailing comma if we are permuting, as simple as that!
and the difference between before and after the modifications is:
sururi@dutsm0175 Octave $ diff genfor.php genfor2.php
10a11,16
> if($_GET["perm"]||$_GET["p"]||$_GET["P"])
> {
> $f_perm = TRUE;
> $K+=1;
> }
>
21c27,30
< fwrite($fp, str_repeat("\t",$m)."printf(\"".str_repeat("%d – ",$M)."%d\\n\",");
—
> if(!$f_perm)fwrite($fp, str_repeat("\t",$m)."printf(\"".str_repeat("%d – ",$M)."%d\\n\",");
> else fwrite($fp, str_repeat("\t",$m)."printf(\"".str_repeat("%d – ",($M-1))."%d\\n\",");
>
> $str = "";
24,25c33,35
< fwrite($fp, "k".$i.",");
< fwrite($fp,($K-1).")\n");
—
> $str .= "k".$i.",";
> if(!$f_perm) fwrite($fp,$str.($K-1).")\n");
> else fwrite($fp, substr($str,0,-1).")\n");
for the really lazy people (like myself), here is the whole deal! 8)
<?
//Support for command line variable passing:
for($i=1;$i<sizeof($_SERVER["argv"]);$i++)
{
list($var0,$val0) = explode("=",$_SERVER["argv"][$i]);
$_GET[$var0] = $val0;
}
$K = $_GET["K"];
$M = $_GET["M"];
if(!$_GET["K"])$_GET["K"] = 10;
if(!$_GET["M"])$_GET["M"] = 2;
if(!$_GET["f"])$_GET["f"] = "lol.m";
if($_GET["perm"]||$_GET["p"]||$_GET["P"])
{
$f_perm = TRUE;
$K+=1;
}
$fp = fopen($_GET["f"],"w");
fwrite($fp, "count=0;\n");
fwrite($fp, str_repeat("\t",$m)."for k0 = 0:".($K-$M-1)."\n");
for($m=1;$m<$M;$m++)
{
fwrite($fp, str_repeat("\t",$m)."for k".$m." = k".($m-1)."+1:".($K-$M-1+$m)."\n");
}
if(!$f_perm)fwrite($fp, str_repeat("\t",$m)."printf(\"".str_repeat("%d – ",$M)."%d\\n\",");
else fwrite($fp, str_repeat("\t",$m)."printf(\"".str_repeat("%d – ",($M-1))."%d\\n\",");
$str = "";
for($i=0;$i<$m;$i++)
$str .= "k".$i.",";
if(!$f_perm) fwrite($fp,$str.($K-1).")\n");
else fwrite($fp, substr($str,0,-1).")\n");
fwrite($fp, str_repeat("\t",$m)."count++;\n");
for($m=$M-1;$m>=0;$m–)
{
fwrite($fp, str_repeat("\t",$m)."endfor\n");
}
fwrite($fp,"if(count!=factorial(".$K."-1)/(factorial(".$K."-".$M."-1)*factorial(".$M.")))\n\tprintf(\"Houston, we have a problem..\");\nendif\n");
fwrite($fp,"printf(\"count : %d\\nfactorial : %d\\n\",count,factorial(".$K."-1)/(factorial(".$K."-".$M."-1)*factorial(".$M.")));\n");
fclose($fp);
system("octave ".$_GET["f"]);
?>
After the modifications,
php genfor.php K=5 M=3 p=1
will produce
0 – 1 – 2
0 – 1 – 3
0 – 1 – 4
0 – 2 – 3
0 – 2 – 4
0 – 3 – 4
1 – 2 – 3
1 – 2 – 4
1 – 3 – 4
2 – 3 – 4
Specification of an extensible and portable file format for electronic structure and crystallographic data
May 6, 2008 Posted by Emre S. Tasci
Specification of an extensible and portable file format for electronic structure and crystallographic data
X. Gonzea, C.-O. Almbladh, A. Cucca, D. Caliste, C. Freysoldt, M.A.L. Marques, V. Olevano, Y. Pouillon and M.J. Verstraet
Comput. Mater. Sci. (2008) doi:10.1016/j.commatsci.2008.02.023
Abstract
In order to allow different software applications, in constant evolution, to interact and exchange data, flexible file formats are needed. A file format specification for different types of content has been elaborated to allow communication of data for the software developed within the European Network of Excellence “NANOQUANTA”, focusing on first-principles calculations of materials and nanosystems. It might be used by other software as well, and is described here in detail. The format relies on the NetCDF binary input/output library, already used in many different scientific communities, that provides flexibility as well as portability across languages and platforms. Thanks to NetCDF, the content can be accessed by keywords, ensuring the file format is extensible and backward compatible.
Keywords
Electronic structure; File format standardization; Crystallographic datafiles; Density datafiles; Wavefunctions datafiles; NetCDF
PACS classification codes
61.68.+n; 63.20.dk; 71.15.Ap
Problems with the binary representations:
- lack of portability between big-endian and little-endian platforms (and vice-versa), or between 32-bit and 64-bit platforms;
- difficulties to read the files written by F77/90 codes from C/C++ software (and vice-versa)
- lack of extensibility, as one file produced for one version of the software might not be readable by a past/forthcoming version
(Network Common Data Form) library solves these issues. (Alas, it is binary and in the examples presented in http://www.unidata.ucar.edu/software/netcdf/examples/files.html, it seems like the CDF representations are larger in size than those of the CDL metadata representations)
The idea of standardization of file formats is not new in the electronic structure community [X. Gonze, G. Zerah, K.W. Jakobsen, and K. Hinsen. Psi-k Newsletter 55, February 2003, pp. 129-134. URL: <http://psi-k.dl.ac.uk>] (This article is a must read article, by the way. It’s a very insightful and concerning overview of the good natured developments in the atomistic/molecular software domain. It is purifying in the attempt to produce something good..). However, it proved difficult to achieve without a formal organization, gathering code developers involved in different software project, with a sufficient incentive to realize effective file exchange between such software.
HDF (Hierarchical Data Format) is also an alternative. NetCDF is simpler to use if the data formats are flat, while HDF has definite advantages if one is dealing with hierarchical formats. Typically, we will need to describe many different multi-dimensional arrays of real or complex numbers, for which NetCDF is an adequate tool.
Although a data specification might be presented irrespective of the library actually used for the implementation, such a freedom might lead to implementations using incompatible file formats (like NetCDF and XML for instance). This possibility would in effect block the expected standardization gain. Thus, as a part of the standardization, we require the future implementations of our specification to rely on the NetCDF library only. (Alternative to independent schemas is presented as a mandatory format which is against the whole idea of development. The reason for NetCDF being incompatible with XML lies solely on the inflexibility/inextensibility of the prior format. Although such a readily defined and built format is adventageous considering the huge numerical data and parameters the ab inito software uses, it is very immobile and unnecessary for compound and element data.)
The compact representation brought by NetCDF can be by-passed in favour of text encoding (very agreable given the usage purposes: XML type extensible schema usage is much more adequate for structure/composition properties). We are aware of several standardization efforts [J. Junquera, M. Verstraete, X. Gonze, unpublished] and [J. Mortensen, F. Jollet, unpublished. Also check Minutes of the discussion on XML format for PAW setups], relying on XML, which emphasize addressing by content to represent such atomic data.
4 types of data types are distinguished:
- (A) The actual numerical data (which defines whether a file contains wavefunctions, a density, etc), for which a name must have been agreed in the specification.
- (B) The auxiliary data that is mandatory to make proper usage of the actual numerical data of A-type. The name and description of this auxiliary information is also agreed.
- (C) The auxiliary data that is not mandatory to make proper usage of the A-type numerical data, but for which a name and description has been agreed in the specification.
- (D) Other data, typically code-dependent, whose availability might help the use of the file for a specific code. The name of these variables should be different from the names chosen for agreed variables of A–C types. Such type D data might even be redundant with type A–C data.
The NetCDF interface adapts the dimension ordering to the programming language used. The notation here is C-like, i.e. row-major storage, the last index varying the fastest. In FORTRAN, a similar memory mapping is obtained by reversing the order of the indices. (So, the ordering/reverse-ordering is handled by the interface/library)
Concluding Remarks
We presented the specifications for a file format relying on the NetCDF I/O library with a content related to electronic structure and crystallographic data. This specification takes advantage of all the interesting properties of NetCDF-based files, in particular portability and extensibility. It is designed for both serial and distributed usage, although the latter characteristics was not presented here.
Several software in the Nanoquanta and ETSF [15] context can produce or read this file format: ABINIT [16] and [17], DP [18], GWST, SELF [19], V_Sim [20]. In order to further encourage its use, a library of Fortran routines [5] has been set up, and is available under the GNU LGPL licence.
Additional Information:
Announcement for a past event
( http://www.tddft.org/pipermail/fsatom/2003-February/000004.html )
CECAM – psi-k – SIMU joint Tutorial
1) Title : Software solutions for data exchange and code gluing.
Location : Lyon Dates : 8-10 october, 2003
Purpose : In this tutorial, we will teach software tools and standards that have recently emerged in view of the exchange of data (text and binary) and gluing of codes : (1) Python, as scripting language, its interfaces with C and FORTRAN ; (2) XML, a standard for representing structured data in text files ; (3) netCDF, a library and file format for the exchange and storage of binary data, and its interfaces with C, Fortran, and Python
Organizers : X. Gonze gonze@pcpm.ucl.ac.be
K. Hinsen hinsen@cnrs-orleans.fr
2) Scientific content
Recent discussions, related to the CECAM workshop on "Open Source Software for Microscopic Simulations", June 19-21, 2002, to the GRID concept (http://www.gridcomputing.com), as well as to the future Integrated Infrastructure Initiative proposal linked to the European psi-k network (http://psi-k.dl.ac.uk), have made clear that one challenge for the coming years is the ability to establish standards for accessing codes, transferring data between codes, testing codes against each other, and become able to "glue" them (this being facilitated by the Free Software concept).
In the present tutorial, we would like to teach three "software solutions" to face this challenge : Python, XML and netCDF.
Python is now the de facto "scripting langage" standard in the computational physics and chemistry community. XML (eXtended Markup Language) is a framework for building mark-up languages, allowing to set-up self-describing documents, readable by humans and machines. netCDF allows binary files to be portable accross platforms. It is not our aim to cover all possible solutions to the above-mentioned challenges (e.g. PERL, Tcl, or HDF), but these three have proven suitable for atomic-scale simulations, in the framework of leading projects like CAMPOS (http://www.fysik.dtu.dk/campos), MMTK (http://dirac.cnrs-orleans.fr/MMTK), and GROMACS (http://www.gromacs.org). Other software projects like ABINIT (http://www.abinit.org) and PWSCF (http://www.pwscf.org – in the DEMOCRITOS context), among others, have made clear their interest for these. All of these software solutions can be used without having to buy a licence.
Tentative program of the tutorial. Lectures in the morning, hands-on training in the afternoon.
1st day ——- 2h Python basics 1h Interface : Python/C or FORTRAN 1h XML basics Afternoon Training with Python, and interfaces with C and FORTRAN
2nd day ——- 2h Python : object oriented (+ an application to GUI and Tk) 1h Interface : Python/XML 1h Interface : XML + C or FORTRAN Afternoon Training with XML + interfaces
3rd day ——- 1h Python : numerical 1h netCDF basics 1h Interface : netCDF/Python 1h Interface : netCDF/C or FORTRAN Afternoon Training with netCDF + interfaces
3) List of lecturers
K. Hinsen (Orleans, France), organizer X. Gonze (Louvain-la-Neuve, Belgium), organizer K. Jakobsen (Lyngby, Denmark), instructor J. Schiotz (Lyngby, Denmark), instructor J. Van Der Spoel (Groningen, The Netherlands), instructor M. van Loewis (Berlin, Germany), instructor
4) Number of participants : around 20, Most of the participants should be PhD students, postdoc or young permanent scientists, involved in code development. It is assumed that the attendants have a good knowledge of UNIX, and C or FORTRAN.
Our budget will allow contributing to travel and local expenses of up to 20 participants.
XML and NetCDF:
from Specification of file formats for NANOQUANTA/ETSF Specification – www.etsf.eu/research/software/nq_specff_v2.1_final.pdf
Section 1. General considerations concerning the present file format specifications.
One has to consider separately the set of data to be included in each of different types of files, from their representation. Concerning the latter, one encounters simple text files, binary files, XML-structured files, NetCDF files, etc … It was already decided previously (Nanoquanta meeting Maratea Sept. 2004) to evolve towards formats that deal appropriately with the self- description issue, i.e. XML and NetCDF. The inherent flexibility of these representations will also allow to evolve specific versions of each type of files progressively, and refine earlier working proposals. The same direction has been adopted by several groups of code developers that we know of.
Information on NetCDF and XML can be obtained from the official Web sites,
http://www.unidata.ucar.edu/software/netcdf/ and
http://www.w3.org/XML/
There are numerous other presentations of these formats on the Web, or in books.
The elaboration of file formats based on NetCDF has advanced a lot during the Louvain-la- Neuve mini-workshop. There has been also some remarks about XML.
Concerning XML :
(A) The XML format is most adapted for the structured representation of relatively small quantity of data, as it is not compressed.
(B) It is a very flexible format, but hard to read in Fortran (no problem in C, C++ or Python). Recently, Alberto Garcia has set up a XMLF90 library of routines to read XML from Fortran. http://lcdx00.wm.lc.ehu.es/~wdpgaara/xml/index.html Other efforts exists in this direction http://nn-online.org/code/xml/
Concerning NetCDF
- (A) Several groups of developers inside NQ have already a good experience of using it, for the representation of binary data (large files).
- (B) Although there is no clear advantage of NetCDF compared to HDF (another possibility for large binary files), this experience inside the NQ network is the main reason for preferring it. By the way, NetCDF and HDF are willing to merge (this might take a few years, though).
- (C) File size limitations of NetCDF exist, see appendix D, but should be overcome in the future.
Thanks to the flexibility of NetCDF, the content of a NetCDF file format suitable for use for NQ softwares might be of four different types :
(1) The actual numerical data (that defines a file for wavefunctions, or a density file, etc …), whose NetCDF description would have been agreed.
(2) The auxiliary data that are mandatory to make proper usage of the actual numerical data. The NetCDF description of these auxiliary data should also be agreed.
(3) The auxiliary data that are not mandatory, but whose NetCDF description has been agreed, in a larger context.
(4) Other data, typically code-dependent, whose existence might help the use of the file for a specific code.
References:
[5] URL: <http://www.etsf.eu/index.php?page=tools>.
[15] URL: <http://www.etsf.eu/>.
[16] X. Gonze, J.-M. Beuken, R. Caracas, F. Detraux, M. Fuchs, G.-M. Rignanese, L. Sindic, M. Verstraete, G. Zerah, F. Jollet, M. Torrent, A. Roy, M. Mikami, Ph. Ghosez, J.-Y. Raty and D.C. Allan, Comput. Mater. Sci. 25 (2002), pp. 478–492.
[17] X. Gonze, G.-M. Rignanese, M. Verstraete, J.-M. Beuken, Y. Pouillon, R. Caracas, F. Jollet, M. Torrent, G. Zérah, M. Mikami, Ph. Ghosez, M. Veithen, J.-Y. Raty, V. Olevano, F. Bruneval, L. Reining, R. Godby, G. Onida, D.R. Hamann and D.C. Allan, Zeit. Kristall. 220 (2005), pp. 558–562.
[18] URL: <http://dp-code.org>.
[19] URL: <http://www.bethe-salpeter.org>.
[20] URL: http://www-drfmc.cea.fr/sp2m/L_Sim/V_Sim/index.en.html.
Element Arrays
April 10, 2008 Posted by Emre S. Tasci
For future reference when coding, I thought it would be useful to include these arrays. All of the arrays are ordered with respect to their Atomic Numbers.
Atomic Numbers Array
"1", "2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12", "13", "14", "15", "16", "17", "18", "19", "20", "21", "22", "23", "24", "25", "26", "27", "28", "29", "30", "31", "32", "33", "34", "35", "36", "37", "38", "39", "40", "41", "42", "43", "44", "45", "46", "47", "48", "49", "50", "51", "52", "53", "54", "55", "56", "57", "58", "59", "60", "61", "62", "63", "64", "65", "66", "67", "68", "69", "70", "71", "72", "73", "74", "75", "76", "77", "78", "79", "80", "81", "82", "83", "84", "85", "86", "87", "88", "89", "90", "91", "92", "93", "94", "95", "96", "97", "98", "99", "100", "101", "102", "103", "104", "105", "106", "107", "108", "109", "110", "111", "112", "113", "114", "115", "116", "117", "118"
Symbols Array
"H", "He", "Li", "Be", "B", "C", "N", "O", "F", "Ne", "Na", "Mg", "Al", "Si", "P", "S", "Cl", "Ar", "K", "Ca", "Sc", "Ti", "V", "Cr", "Mn", "Fe", "Co", "Ni", "Cu", "Zn", "Ga", "Ge", "As", "Se", "Br", "Kr", "Rb", "Sr", "Y", "Zr", "Nb", "Mo", "Tc", "Ru", "Rh", "Pd", "Ag", "Cd", "In", "Sn", "Sb", "Te", "I", "Xe", "Cs", "Ba", "La", "Ce", "Pr", "Nd", "Pm", "Sm", "Eu", "Gd", "Tb", "Dy", "Ho", "Er", "Tm", "Yb", "Lu", "Hf", "Ta", "W", "Re", "Os", "Ir", "Pt", "Au", "Hg", "Tl", "Pb", "Bi", "Po", "At", "Rn", "Fr", "Ra", "Ac", "Th", "Pa", "U", "Np", "Pu", "Am", "Cm", "Bk", "Cf", "Es", "Fm", "Md", "No", "Lr", "Rf", "Db", "Sg", "Bh", "Hs", "Mt", "Ds", "Rg", "Uub", "Uut", "Uuq", "Uup", "Uuh", "Uus", "Uuo"
Names Array
"Hydrogen", "Helium", "Lithium", "Beryllium", "Boron", "Carbon", "Nitrogen", "Oxygen", "Fluorine", "Neon", "Sodium", "Magnesium", "Aluminum", "Silicon", "Phosphorus", "Sulfur", "Chlorine", "Argon", "Potassium", "Calcium", "Scandium", "Titanium", "Vanadium", "Chromium", "Manganese", "Iron", "Cobalt", "Nickel", "Copper", "Zinc", "Gallium", "Germanium", "Arsenic", "Selenium", "Bromine", "Krypton", "Rubidium", "Strontium", "Yttrium", "Zirconium", "Niobium", "Molybdenum", "Technetium", "Ruthenium", "Rhodium", "Palladium", "Silver", "Cadmium", "Indium", "Tin", "Antimony", "Tellurium", "Iodine", "Xenon", "Cesium", "Barium", "Lanthanum", "Cerium", "Praseodymium", "Neodymium", "Promethium", "Samarium", "Europium", "Gadolinium", "Terbium", "Dysprosium", "Holmium", "Erbium", "Thulium", "Ytterbium", "Lutetium", "Hafnium", "Tantalum", "Tungsten", "Rhenium", "Osmium", "Iridium", "Platinum", "Gold", "Mercury", "Thallium", "Lead", "Bismuth", "Polonium", "Astatine", "Radon", "Francium", "Radium", "Actinium", "Thorium", "Protactinium", "Uranium", "Neptunium", "Plutonium", "Americium", "Curium", "Berkelium", "Californium", "Einsteinium", "Fermium", "Mendelevium", "Nobelium", "Lawrencium", "Rutherfordium", "Dubnium", "Seaborgium", "Bohrium", "Hassium", "Meitnerium", "Darmstadtium", "Roentgenium", "Ununbium", "Ununtrium", "Ununquadium", "Ununpentium", "Ununhexium", "Ununseptium", "Ununoctium"
Pettifor’s Mendeleev Numbers Array
D.G. Pettifor, Mater. Sci. Tech. 4 (1988) 2480 | D.G. Pettifor, Bonding and Structure of Molecules and Solids, Oxford University Press (1995) p.13
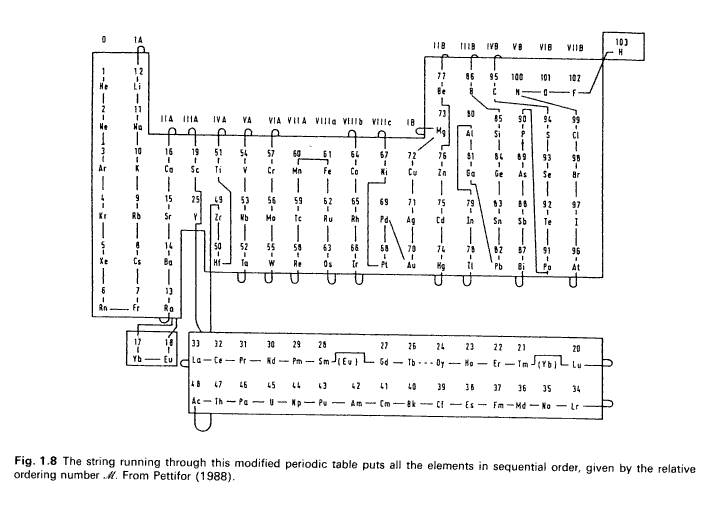
"103", "1", "12", "77", "86", "95", "100", "101", "102", "2", "11", "73", "80", "85", "90", "94", "99", "3", "10", "16", "19", "51", "54", "57", "60", "61", "64", "67", "72", "76", "81", "84", "89", "93", "98", "4", "9", "15", "25", "49", "53", "56", "59", "62", "65", "69", "71", "75", "79", "83", "88", "92", "97", "5", "8", "14", "33", "32", "31", "30", "29", "28", "18", "27", "26", "24", "23", "22", "21", "17", "20", "50", "52", "55", "58", "63", "66", "68", "70", "74", "78", "82", "87", "91", "96", "6", "7", "13", "48", "47", "46", "45", "44", "43", "42", "41", "40", "39", "38", "37", "36", "35", "34", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0"
Villars’ Mendeleev Numbers Array
P. Villars et al., J. Alloys Compd. 367 (2004) 167] – ![]() doi:10.1016/j.jallcom.2003.08.060
doi:10.1016/j.jallcom.2003.08.060

"92", "98", "1", "67", "72", "77", "82", "87", "93", "99", "2", "68", "73", "78", "83", "88", "94", "100", "3", "7", "11", "43", "46", "49", "52", "55", "58", "61", "64", "69", "74", "79", "84", "89", "95", "101", "4", "8", "12", "44", "47", "50", "53", "56", "59", "62", "65", "70", "75", "80", "85", "90", "96", "102", "5", "9", "13", "15", "17", "19", "21", "23", "25", "27", "29", "31", "33", "35", "37", "39", "41", "45", "48", "51", "54", "57", "60", "63", "66", "71", "76", "81", "86", "91", "97", "103", "6", "10", "14", "16", "18", "20", "22", "24", "26", "28", "30", "32", "34", "36", "38", "40", "42", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0"
You can also download the SQL version for these values from here.
Hope it helps..


